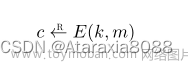来源:《斯坦福数据挖掘教程·第三版》对应的公开英文书和PPT
Chapter 11 Dimensionality Reduction
Let M be a square matrix. Let λ be a constant and e a nonzero column vector with the same number of rows as M. Then λ is an eigenvalue of M and e is the corresponding eigenvector of M if M e = λ e Me = λe Me=λe.
Start with any unit vector v of the appropriate length and compute
M
i
v
M^iv
Miv iteratively until it converges. When M is a stochastic matrix, the limiting vector is the principal eigenvector (the eigenvector with the largest eigenvalue), and its corresponding eigenvalue is 1. This method for finding the principal eigenvector, called power iteration, works quite generally, although if the principal eigenvalue (eigenvalue associated with the principal eigenvector) is not 1, then as i grows, the ratio of
M
i
+
1
v
M^{i+1}v
Mi+1v to
M
i
v
M^iv
Miv approaches the principal eigenvalue while
M
i
v
M^iv
Miv approaches
a vector (probably not a unit vector) with the same direction as the principal eigenvector.
To find the second eigenpair we create a new matrix M ∗ = M − λ 1 x x T M^∗ = M − λ_1xx^T M∗=M−λ1xxT. Then, use power iteration on M ∗ M^∗ M∗ to compute its largest eigenvalue. The obtained x ∗ x^∗ x∗ and λ ∗ λ^∗ λ∗ correspond to the second largest eigenvalue and the corresponding eigenvector of matrix M. Intuitively, what we have done is eliminate the influence of a given eigenvector by setting its associated eigenvalue to zero. The formal justification is the following two observations. If M ∗ = M − λ x x T M^∗ = M − λxx^T M∗=M−λxxT, where x and λ are the eigenpair with the largest eigenvalue, then:
-
x is also an eigenvector of M ∗ M^∗ M∗, and its corresponding eigenvalue is 0. In proof, observe that
M ∗ x = ( M − λ x x T ) x = M x − λ x x T x = M x − λ x = 0 M^∗x = (M − λxx^T)x = Mx − λxx^Tx = Mx − λx = 0 M∗x=(M−λxxT)x=Mx−λxxTx=Mx−λx=0
At the next-to-last step we use the fact that x T x = 1 x^Tx = 1 xTx=1 because x is a unit vector.
-
Conversely, if v and λ v λ_v λv are an eigenpair of a symmetric matrix M other than the first eigenpair (x, λ), then they are also an eigenpair of M ∗ M^∗ M∗.
Proof :M ∗ v = ( M ∗ ) T v = ( M − λ x x T ) T v = M T v − λ x ( x T v ) = M T v = λ v v M^∗v = (M^∗)^Tv = (M − λxx^T)^Tv = M^Tv − λx(x^Tv) = M^Tv = λ_vv M∗v=(M∗)Tv=(M−λxxT)Tv=MTv−λx(xTv)=MTv=λvv
This sequence of equalities needs the following justifications:
(a) If M is symmetric, then M = M T M = M^T M=MT.
(b) The eigenvectors of a symmetric matrix are orthogonal. That is, the dot product of any two distinct eigenvectors of a matrix is 0. We do not prove this statement here.
Principal-component analysis, or PCA, is a technique for taking a dataset consisting of a set of tuples representing points in a high-dimensional space and finding the directions along which the tuples line up best. The idea is to treat the set of tuples as a matrix M and find the eigenvectors for M M T MM^T MMT or M T M M^TM MTM. The matrix of these eigenvectors can be thought of as a rigid rotation in a high dimensional space. When you apply this transformation to the original data, the axis corresponding to the principal eigenvector is the one along which the points are most “spread out,” More precisely, this axis is the one along which the variance of the data is maximized. Put another way, the points can best be viewed as lying along this axis, with small deviations from this axis. Likewise, the axis corresponding to the second eigenvector (the eigenvector corresponding to the second-largest eigenvalue) is the axis along which the variance of distances from the first axis is greatest, and so on.
Any matrix of orthonormal vectors (unit vectors that are orthogonal to one another) represents a rotation and/or reflection of the axes of a Euclidean space.
We conclude that the eigenvalues of M M T MM^T MMT are the eigenvalues of M T M M^TM MTM plus additional 0’s. If the dimension of M M T MM^T MMT were less than the dimension off M T M M^TM MTM, then the opposite would be true; the eigenvalues of M T M M^TM MTM would be those of M M T MM^T MMT plus additional 0’s.
Let M be an m × n m × n m×n matrix, and let the rank of M be r. Recall that the rank of a matrix is the largest number of rows (or equivalently columns) we can choose for which no nonzero linear combination of the rows is the all-zero vector 0 (we say a set of such rows or columns is independent). Then we can find matrices U, Σ, and V as shown in Fig. 11.5 with the following properties:
- U is an m × r m × r m×r column-orthonormal matrix; that is, each of its columns is a unit vector and the dot product of any two columns is 0.
- V is an n × r n × r n×r column-orthonormal matrix. Note that we always use V in its transposed form, so it is the rows of V T V^T VT that are orthonormal.
- Σ is a diagonal matrix; that is, all elements not on the main diagonal are 0. The elements of Σ are called the singular values of M.

Suppose we want to represent a very large matrix M by its SVD components U, Σ, and V , but these matrices are also too large to store conveniently. The best way to reduce the dimensionality of the three matrices is to set the smallest of the singular values to zero. If we set the s smallest singular values to 0, then we can also eliminate the corresponding s columns of U and V.
How Many Singular Values Should We Retain?
A useful rule of thumb is to retain enough singular values to make up 90% of the energy in Σ. That is, the sum of the squares of the retained singular values should be at least 90% of the sum of the squares of all the singular values.
The choice of the lowest singular values to drop when we reduce the number of dimensions can be shown to minimize the root-mean-square error between the original matrix M and its approximation.
It says that V is the matrix of eigenvectors of M T M M^TM MTM and Σ 2 Σ^2 Σ2 is the diagonal matrix whose entries are the corresponding eigenvalues.
Thus, the same algorithm that computes the eigenpairs for M T M M^TM MTM gives us the matrix V for the SVD of M itself. It also gives us the singular values for this SVD; just take the square roots of the eigenvalues for M T M M^TM MTM. U is the matrix of eigenvectors of M M T MM^T MMT.
Definition of CUR
Let M be a matrix of m rows and n columns. Pick a target number of “concepts” r to be used in the decomposition. A CUR-decomposition of M is a randomly chosen set of r columns of M, which form the m × r m × r m×r matrix C, and a randomly chosen set of r rows of M, which form the r × n r × n r×n matrix R. There is also an r × r r × r r×r matrix U that is constructed from C and R as follows:
- Let W be the r × r r × r r×r matrix that is the intersection of the chosen columns of C and the chosen rows of R. That is, the element in row i and column j of W is the element of M whose column is the jth column of C and whose row is the ith row of R.
- Compute the SVD of W; say W = X Σ Y T W = XΣY^T W=XΣYT.
- Compute Σ + Σ^+ Σ+, the Moore-Penrose pseudoinverse of the diagonal matrix Σ. That is, if the ith diagonal element of Σ is σ ≠ 0 σ \ne 0 σ=0, then replace it by 1/σ. But if the ith element is 0, leave it as 0.
- Let U = Y ( Σ + ) 2 X T U = Y (Σ^+)^2X^T U=Y(Σ+)2XT.
Having selected each of the columns of M, we scale each column by dividing its elements by the square root of the expected number of times this column would be picked. That is, we divide the elements of the jth column of M, if it is selected, by
r
q
j
\sqrt {rq_j}
rqj . The scaled column of M becomes a column of C.
Rows of M are selected for R in the analogous way. For each row of R we select from the rows of M, choosing row i with probability
p
i
p_i
pi. Recall
p
i
p_i
pi is the sum of the squares of the elements of the ith row divided by the sum of the squares of all the elements of M. We then scale each chosen row by dividing by
r
p
i
\sqrt {rp_i}
rpi if it is the ith row of M that was chosen.
It is quite possible that a single row or column is selected more than once. However, it is also possible to combine k rows of R that are each the same row of the matrix M into a single row of R, thus leaving R with fewer rows. Likewise, k columns of C that each come from the same column of M can be combined into one column of C. However, for either rows or columns,
the remaining vector should have each of its elements multiplied by
k
\sqrt k
k .
When we merge some rows and/or columns, it is possible that R has fewer rows than C has columns, or vice versa. As a consequence, W will not be a square matrix. However, we can still take its pseudoinverse by decomposing it into
W
=
X
Σ
Y
T
W = XΣY^T
W=XΣYT, where Σ is now a diagonal matrix with some all-0 rows or columns, whichever it has more of. To take the pseudoinverse of such a diagonal matrix, we treat each element on the diagonal as usual (invert nonzero elements
and leave 0 as it is), but then we must transpose the result.文章来源:https://www.toymoban.com/news/detail-469497.html
Summary of Chapter 11
- Dimensionality Reduction: The goal of dimensionality reduction is to replace a large matrix by two or more other matrices whose sizes are much smaller than the original, but from which the original can be approximately reconstructed, usually by taking their product.
- Eigenvalues and Eigenvectors: A matrix may have several eigenvectors such that when the matrix multiplies the eigenvector, the result is a constant multiple of the eigenvector. That constant is the eigenvalue associated with this eigenvector. Together the eigenvector and its eigenvalue are called an eigenpair.
- Finding Eigenpairs by Power Iteration: We can find the principal eigenvector (eigenvector with the largest eigenvalue) by starting with any vector and repeatedly multiplying the current vector by the matrix to get a new vector. When the changes to the vector become small, we can treat the result as a close approximation to the principal eigenvector. By modifying the matrix, we can then use the same iteration to get the second eigenpair (that with the second-largest eigenvalue), and similarly get each of the eigenpairs in turn, in order of decreasing value of the eigenvalue.
- Principal-Component Analysis: This technique for dimensionality reduction views data consisting of a collection of points in a multidimensional space as a matrix, with rows corresponding to the points and columns to the dimensions. The product of this matrix and its transpose has eigenpairs, and the principal eigenvector can be viewed as the direction in the space along which the points best line up. The second eigenvector represents the direction in which deviations from the principal eigenvector are the greatest, and so on.
- Dimensionality Reduction by PCA: By representing the matrix of points by a small number of its eigenvectors, we can approximate the data in a way that minimizes the root-mean-square error for the given number of columns in the representing matrix.
- Singular-Value Decomposition: The singular-value decomposition of a matrix consists of three matrices, U, Σ, and V . The matrices U and V are column-orthonormal, meaning that as vectors, the columns are orthogonal, and their lengths are 1. The matrix Σ is a diagonal matrix, and the values along its diagonal are called singular values. The product of U, Σ, and the transpose of V equals the original matrix.
- Concepts: SVD is useful when there are a small number of concepts that connect the rows and columns of the original matrix. For example, if the original matrix represents the ratings given by movie viewers (rows) to movies (columns), the concepts might be the genres of the movies. The matrix U connects rows to concepts, Σ represents the strengths of the concepts, and V connects the concepts to columns.
- Queries Using the Singular-Value Decomposition: We can use the decomposition to relate new or hypothetical rows of the original matrix to the concepts represented by the decomposition. Multiply a row by the matrix V of the decomposition to get a vector indicating the extent to which that row matches each of the concepts.
- Using SVD for Dimensionality Reduction: In a complete SVD for a matrix, U and V are typically as large as the original. To use fewer columns for U and V , delete the columns corresponding to the smallest singular values from U, V , and Σ. This choice minimizes the error in reconstructing the original matrix from the modified U, Σ, and V .
- Decomposing Sparse Matrices: Even in the common case where the given matrix is sparse, the matrices constructed by SVD are dense. The CUR decomposition seeks to decompose a sparse matrix into sparse, smaller matrices whose product approximates the original matrix.
-
CUR Decomposition: This method chooses from a given sparse matrix a set of columns C and a set of rows R, which play the role of U and
V
T
V^T
VT in SVD; the user can pick any number of rows and columns. The choice of rows and columns is made randomly with a distribution that depends on the Frobenius norm, or the square root of the sum of the
squares of the elements. Between C and R is a square matrix called U that is constructed by a pseudo-inverse of the intersection of the chosen rows and columns.
END文章来源地址https://www.toymoban.com/news/detail-469497.html
到了这里,关于《斯坦福数据挖掘教程·第三版》读书笔记(英文版)Chapter 11 Dimensionality Reduction的文章就介绍完了。如果您还想了解更多内容,请在右上角搜索TOY模板网以前的文章或继续浏览下面的相关文章,希望大家以后多多支持TOY模板网!