[论文笔记] Swin UNETR 论文笔记: MRI 图像脑肿瘤语义分割
Author: Sijin Yu
[1] Ali Hatamizadeh, Vishwesh Nath, Yucheng Tang, Dong Yang, Holger R. Roth, and Daguang Xu. Swin UNETR: Swin Transformers for Semantic Segmentation of Brain Tumors in MRI Images. MICCAI, 2022.
📎开源代码链接
1. Abstract
- 脑肿瘤的语义分割是一项基本的医学影像分析任务, 涉及多种 MRI 成像模态, 可协助临床医生诊断病人并随后研究恶性实体的进展.
- 近年来, 完全卷积神经网络 (Fully Convolutional Neural Networks, FCNNs) 方法已成为 3D 医学影像分割的事实标准.
- 流行的 “U形” 网络架构在不同的 2D 和 3D 语义分割任务以及各种成像模式上实现了最先进的性能基准.
- 然而, 由于 FCNNs 中卷积层的核大小有限, 它们在建模长距离信息方面的性能是次优的, 这可能导致在分割大小不一的肿瘤时出现缺陷.
- 另一方面, Transformer 模型在多个领域展示了捕获长距离信息的卓越能力, 包括自然语言处理和计算机视觉.
- 受 ViT 及其变体成功的启发, 我们提出了一种名为 Swin UNEt TRansformers (Swin UNETR) 的新型分割模型.
- 具体来说, 3D 脑肿瘤语义分割任务被重新定义为序列到序列预测问题, 其中多模态输入数据被投影成一维嵌入序列, 并用作层级 Swin 变换器编码器的输入.
- Swin Transformer 编码器使用移位窗口计算自注意力, 在五个不同的分辨率上提取特征, 并通过跳跃连接在每个分辨率上连接到基于FCNN 的解码器.
- 我们参加了 2021 年 BraTS 分割挑战赛, 我们提出的模型在验证阶段位列表现最佳的方法之一.
2. Motivation & Contribution
2.1 Motivation
- 在医疗保健的人工智能领域, 特别是脑肿瘤分析中, 需要更先进的分割技术来准确划定肿瘤, 以便诊断和术前规划.
- 当前基于 CNN 的脑肿瘤分割方法由于其小感受野, 难以捕捉长距离依赖关系.
- ViTs 在捕捉各种领域的长距离信息方面显示出潜力, 暗示其在改善医学图像分割中的适用性.
2.2 Contribution
- 提出了一种新型架构, Swin UNEt TRansformers (Swin UNETR), 结合了 Swin Transformer 编码器与 U 形 CNN 解码器, 用于多模态三维脑肿瘤分割.
- 在 2021 年多模态脑肿瘤分割挑战 (BraTS) 中展示了 Swin UNETR 模型的有效性, 验证阶段取得了排名靠前的成绩, 并在测试中表现出竞争力.
3. Model
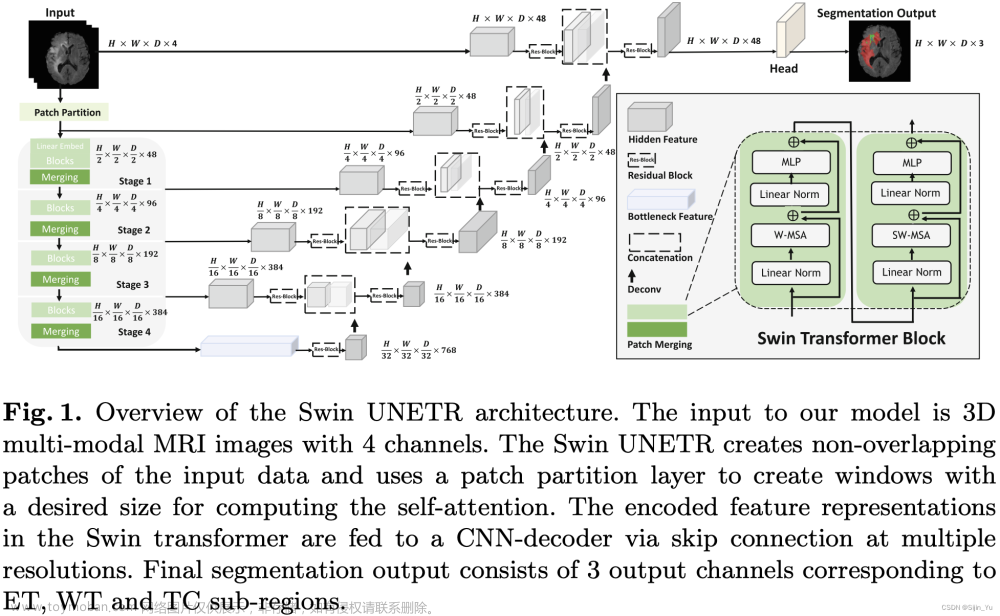
-
将输入的图像打成 Patch.
输入的图像为 X ∈ R H × W × D × S X\in\mathbb R^{H\times W\times D\times S} X∈RH×W×D×S. 一个 Patch 的分辨率为 ( H ′ , W ′ , D ′ ) (H',W',D') (H′,W′,D′), 一个 Patch 的形状为 R H ′ × W ′ × D ′ × S \mathbb R^{H'\times W'\times D'\times S} RH′×W′×D′×S.
则图像变为一个 Patch 的序列, 序列长度为 ⌈ H H ′ ⌉ × ⌈ W W ′ ⌉ × ⌈ D D ′ ⌉ \lceil\frac{H}{H'}\rceil\times\lceil\frac{W}{W'}\rceil\times\lceil\frac{D}{D'}\rceil ⌈H′H⌉×⌈W′W⌉×⌈D′D⌉.
在本文中, Patch size 为 ( H ′ , W ′ , D ′ ) = ( 2 , 2 , 2 ) (H',W',D')=(2, 2, 2) (H′,W′,D′)=(2,2,2).
对于每个 patch, 将其映射为一个嵌入维度为 C C C 的 token. 因此, 最终得到分辨率为 ( ⌈ H H ′ ⌉ , ⌈ W W ′ ⌉ , ⌈ D D ′ ⌉ ) (\lceil\frac{H}{H'}\rceil,\lceil\frac{W}{W'}\rceil,\lceil\frac{D}{D'}\rceil) (⌈H′H⌉,⌈W′W⌉,⌈D′D⌉) 的 3D tokens.
-
对 3D tokens 应用 Swin Transformer.
一层 Swin Transformer Block 由两个子层组成: W-MSA, SW-MSA.
经过一层 Swin Transformer Block, 一个 3D tokens 每个方向上的分辨率变为原来的 1 2 \frac12 21, 通道数变为原来的 2 2 2 倍. 见 Fig.1 的左下角.
W-MSA 和 SW-MSA 分别是规则的、循环移动的 partitioning multi-head self-attention, 如下图所示.

4. Experiment
4.1 Dataset
- BraTS 2021
4.2 对比实验

5. Code
以下链接提供了使用Swin UNETR模型进行BraTS21脑肿瘤分割的教程:
下面是部分核心代码注释:文章来源:https://www.toymoban.com/news/detail-851399.html
5.1 数据预处理和增强
from monai import transforms
train_transform = transforms.Compose(
[
# 读入图像
transforms.LoadImaged(keys=["image", "label"]),
# 将单通道的标签图像转换成多通道格式, 每个通道表示不同的肿瘤类别. (转换前是所有类别标签图共用一个单通道图像) transforms.ConvertToMultiChannelBasedOnBratsClassesd(keys="label"),
# 裁剪掉图像周围的背景区域
transforms.CropForegroundd(
keys=["image", "label"],
source_key="image",
k_divisible=[roi[0], roi[1], roi[2]],
),
# 将图像随机裁剪为指定大小
transforms.RandSpatialCropd(
keys=["image", "label"],
roi_size=[roi[0], roi[1], roi[2]],
random_size=False,
),
# 在0轴方向上随机翻转
transforms.RandFlipd(keys=["image", "label"], prob=0.5, spatial_axis=0),
# 在1轴方向上随机翻转
transforms.RandFlipd(keys=["image", "label"], prob=0.5, spatial_axis=1),
# 在2轴方向上随机翻转
transforms.RandFlipd(keys=["image", "label"], prob=0.5, spatial_axis=2),
# 对每个单独通道, 进行强度归一化, 且忽略0值
transforms.NormalizeIntensityd(keys="image", nonzero=True, channel_wise=True),
# 随机调整图像的强度, img = img * (1 + eps)
transforms.RandScaleIntensityd(keys="image", factors=0.1, prob=1.0),
# 随机调整图像的强度, img = img + eps
transforms.RandShiftIntensityd(keys="image", offsets=0.1, prob=1.0),
]
)
val_transform = transforms.Compose(
[
transforms.LoadImaged(keys=["image", "label"]),
transforms.ConvertToMultiChannelBasedOnBratsClassesd(keys="label"),
transforms.NormalizeIntensityd(keys="image", nonzero=True, channel_wise=True),
]
)
5.2 Swin UNETR 模型架构
def forward(self, x_in):
if not torch.jit.is_scripting():
self._check_input_size(x_in.shape[2:])
hidden_states_out = self.swinViT(x_in, self.normalize)
enc0 = self.encoder1(x_in)
enc1 = self.encoder2(hidden_states_out[0])
enc2 = self.encoder3(hidden_states_out[1])
enc3 = self.encoder4(hidden_states_out[2])
dec4 = self.encoder10(hidden_states_out[4])
dec3 = self.decoder5(dec4, hidden_states_out[3])
dec2 = self.decoder4(dec3, enc3)
dec1 = self.decoder3(dec2, enc2)
dec0 = self.decoder2(dec1, enc1)
out = self.decoder1(dec0, enc0)
logits = self.out(out)
return logits
组件的定义如下:文章来源地址https://www.toymoban.com/news/detail-851399.html
self.normalize = normalize
self.swinViT = SwinTransformer(
in_chans=in_channels,
embed_dim=feature_size,
window_size=window_size,
patch_size=patch_sizes,
depths=depths,
num_heads=num_heads,
mlp_ratio=4.0,
qkv_bias=True,
drop_rate=drop_rate,
attn_drop_rate=attn_drop_rate,
drop_path_rate=dropout_path_rate,
norm_layer=nn.LayerNorm,
use_checkpoint=use_checkpoint,
spatial_dims=spatial_dims,
downsample=look_up_option(downsample, MERGING_MODE) if isinstance(downsample, str) else downsample,
use_v2=use_v2,
)
self.encoder1 = UnetrBasicBlock(
spatial_dims=spatial_dims,
in_channels=in_channels,
out_channels=feature_size,
kernel_size=3,
stride=1,
norm_name=norm_name,
res_block=True,
)
self.encoder2 = UnetrBasicBlock(
spatial_dims=spatial_dims,
in_channels=feature_size,
out_channels=feature_size,
kernel_size=3,
stride=1,
norm_name=norm_name,
res_block=True,
)
self.encoder3 = UnetrBasicBlock(
spatial_dims=spatial_dims,
in_channels=2 * feature_size,
out_channels=2 * feature_size,
kernel_size=3,
stride=1,
norm_name=norm_name,
res_block=True,
)
self.encoder4 = UnetrBasicBlock(
spatial_dims=spatial_dims,
in_channels=4 * feature_size,
out_channels=4 * feature_size,
kernel_size=3,
stride=1,
norm_name=norm_name,
res_block=True,
)
self.encoder10 = UnetrBasicBlock(
spatial_dims=spatial_dims,
in_channels=16 * feature_size,
out_channels=16 * feature_size,
kernel_size=3,
stride=1,
norm_name=norm_name,
res_block=True,
)
self.decoder5 = UnetrUpBlock(
spatial_dims=spatial_dims,
in_channels=16 * feature_size,
out_channels=8 * feature_size,
kernel_size=3,
upsample_kernel_size=2,
norm_name=norm_name,
res_block=True,
)
self.decoder4 = UnetrUpBlock(
spatial_dims=spatial_dims,
in_channels=feature_size * 8,
out_channels=feature_size * 4,
kernel_size=3,
upsample_kernel_size=2,
norm_name=norm_name,
res_block=True,
)
self.decoder3 = UnetrUpBlock(
spatial_dims=spatial_dims,
in_channels=feature_size * 4,
out_channels=feature_size * 2,
kernel_size=3,
upsample_kernel_size=2,
norm_name=norm_name,
res_block=True,
)
self.decoder2 = UnetrUpBlock(
spatial_dims=spatial_dims,
in_channels=feature_size * 2,
out_channels=feature_size,
kernel_size=3,
upsample_kernel_size=2,
norm_name=norm_name,
res_block=True,
)
self.decoder1 = UnetrUpBlock(
spatial_dims=spatial_dims,
in_channels=feature_size,
out_channels=feature_size,
kernel_size=3,
upsample_kernel_size=2,
norm_name=norm_name,
res_block=True,
)
self.out = UnetOutBlock(spatial_dims=spatial_dims, in_channels=feature_size, out_channels=out_channels)
5.2.1 SwinTransformer
class SwinTransformer(nn.Module):
"""
Swin Transformer based on: "Liu et al.,
Swin Transformer: Hierarchical Vision Transformer using Shifted Windows
<https://arxiv.org/abs/2103.14030>"
https://github.com/microsoft/Swin-Transformer
"""
def __init__(
self,
in_chans: int,
embed_dim: int,
window_size: Sequence[int],
patch_size: Sequence[int],
depths: Sequence[int],
num_heads: Sequence[int],
mlp_ratio: float = 4.0,
qkv_bias: bool = True,
drop_rate: float = 0.0,
attn_drop_rate: float = 0.0,
drop_path_rate: float = 0.0,
norm_layer: type[LayerNorm] = nn.LayerNorm,
patch_norm: bool = False,
use_checkpoint: bool = False,
spatial_dims: int = 3,
downsample="merging",
use_v2=False,
) -> None:
"""
Args:
in_chans: dimension of input channels.
embed_dim: number of linear projection output channels.
window_size: local window size.
patch_size: patch size.
depths: number of layers in each stage.
num_heads: number of attention heads.
mlp_ratio: ratio of mlp hidden dim to embedding dim.
qkv_bias: add a learnable bias to query, key, value.
drop_rate: dropout rate.
attn_drop_rate: attention dropout rate.
drop_path_rate: stochastic depth rate.
norm_layer: normalization layer.
patch_norm: add normalization after patch embedding.
use_checkpoint: use gradient checkpointing for reduced memory usage.
spatial_dims: spatial dimension.
downsample: module used for downsampling, available options are `"mergingv2"`, `"merging"` and a
user-specified `nn.Module` following the API defined in :py:class:`monai.networks.nets.PatchMerging`.
The default is currently `"merging"` (the original version defined in v0.9.0).
use_v2: using swinunetr_v2, which adds a residual convolution block at the beginning of each swin stage.
"""
super().__init__()
self.num_layers = len(depths)
self.embed_dim = embed_dim
self.patch_norm = patch_norm
self.window_size = window_size
self.patch_size = patch_size
self.patch_embed = PatchEmbed(
patch_size=self.patch_size,
in_chans=in_chans,
embed_dim=embed_dim,
norm_layer=norm_layer if self.patch_norm else None, # type: ignore
spatial_dims=spatial_dims,
)
self.pos_drop = nn.Dropout(p=drop_rate)
dpr = [x.item() for x in torch.linspace(0, drop_path_rate, sum(depths))]
self.use_v2 = use_v2
self.layers1 = nn.ModuleList()
self.layers2 = nn.ModuleList()
self.layers3 = nn.ModuleList()
self.layers4 = nn.ModuleList()
if self.use_v2:
self.layers1c = nn.ModuleList()
self.layers2c = nn.ModuleList()
self.layers3c = nn.ModuleList()
self.layers4c = nn.ModuleList()
down_sample_mod = look_up_option(downsample, MERGING_MODE) if isinstance(downsample, str) else downsample
for i_layer in range(self.num_layers):
layer = BasicLayer(
dim=int(embed_dim * 2**i_layer),
depth=depths[i_layer],
num_heads=num_heads[i_layer],
window_size=self.window_size,
drop_path=dpr[sum(depths[:i_layer]) : sum(depths[: i_layer + 1])],
mlp_ratio=mlp_ratio,
qkv_bias=qkv_bias,
drop=drop_rate,
attn_drop=attn_drop_rate,
norm_layer=norm_layer,
downsample=down_sample_mod,
use_checkpoint=use_checkpoint,
)
if i_layer == 0:
self.layers1.append(layer)
elif i_layer == 1:
self.layers2.append(layer)
elif i_layer == 2:
self.layers3.append(layer)
elif i_layer == 3:
self.layers4.append(layer)
if self.use_v2:
layerc = UnetrBasicBlock(
spatial_dims=3,
in_channels=embed_dim * 2**i_layer,
out_channels=embed_dim * 2**i_layer,
kernel_size=3,
stride=1,
norm_name="instance",
res_block=True,
)
if i_layer == 0:
self.layers1c.append(layerc)
elif i_layer == 1:
self.layers2c.append(layerc)
elif i_layer == 2:
self.layers3c.append(layerc)
elif i_layer == 3:
self.layers4c.append(layerc)
self.num_features = int(embed_dim * 2 ** (self.num_layers - 1))
def proj_out(self, x, normalize=False):
if normalize:
x_shape = x.size()
if len(x_shape) == 5:
n, ch, d, h, w = x_shape
x = rearrange(x, "n c d h w -> n d h w c")
x = F.layer_norm(x, [ch])
x = rearrange(x, "n d h w c -> n c d h w")
elif len(x_shape) == 4:
n, ch, h, w = x_shape
x = rearrange(x, "n c h w -> n h w c")
x = F.layer_norm(x, [ch])
x = rearrange(x, "n h w c -> n c h w")
return x
def forward(self, x, normalize=True):
x0 = self.patch_embed(x)
x0 = self.pos_drop(x0)
x0_out = self.proj_out(x0, normalize)
if self.use_v2:
x0 = self.layers1c[0](x0.contiguous())
x1 = self.layers1[0](x0.contiguous())
x1_out = self.proj_out(x1, normalize)
if self.use_v2:
x1 = self.layers2c[0](x1.contiguous())
x2 = self.layers2[0](x1.contiguous())
x2_out = self.proj_out(x2, normalize)
if self.use_v2:
x2 = self.layers3c[0](x2.contiguous())
x3 = self.layers3[0](x2.contiguous())
x3_out = self.proj_out(x3, normalize)
if self.use_v2:
x3 = self.layers4c[0](x3.contiguous())
x4 = self.layers4[0](x3.contiguous())
x4_out = self.proj_out(x4, normalize)
return [x0_out, x1_out, x2_out, x3_out, x4_out]
5.2.2 UnetrBasicBlock
class UnetrBasicBlock(nn.Module):
"""
A CNN module that can be used for UNETR, based on: "Hatamizadeh et al.,
UNETR: Transformers for 3D Medical Image Segmentation <https://arxiv.org/abs/2103.10504>"
"""
def __init__(
self,
spatial_dims: int,
in_channels: int,
out_channels: int,
kernel_size: Sequence[int] | int,
stride: Sequence[int] | int,
norm_name: tuple | str,
res_block: bool = False,
) -> None:
"""
Args:
spatial_dims: number of spatial dimensions.
in_channels: number of input channels.
out_channels: number of output channels.
kernel_size: convolution kernel size.
stride: convolution stride.
norm_name: feature normalization type and arguments.
res_block: bool argument to determine if residual block is used.
"""
super().__init__()
if res_block:
self.layer = UnetResBlock(
spatial_dims=spatial_dims,
in_channels=in_channels,
out_channels=out_channels,
kernel_size=kernel_size,
stride=stride,
norm_name=norm_name,
)
else:
self.layer = UnetBasicBlock( # type: ignore
spatial_dims=spatial_dims,
in_channels=in_channels,
out_channels=out_channels,
kernel_size=kernel_size,
stride=stride,
norm_name=norm_name,
)
def forward(self, inp):
return self.layer(inp)
5.2.3 UnetrUpBlock
class UnetrUpBlock(nn.Module):
"""
An upsampling module that can be used for UNETR: "Hatamizadeh et al.,
UNETR: Transformers for 3D Medical Image Segmentation <https://arxiv.org/abs/2103.10504>"
"""
def __init__(
self,
spatial_dims: int,
in_channels: int,
out_channels: int,
kernel_size: Sequence[int] | int,
upsample_kernel_size: Sequence[int] | int,
norm_name: tuple | str,
res_block: bool = False,
) -> None:
"""
Args:
spatial_dims: number of spatial dimensions.
in_channels: number of input channels.
out_channels: number of output channels.
kernel_size: convolution kernel size.
upsample_kernel_size: convolution kernel size for transposed convolution layers.
norm_name: feature normalization type and arguments.
res_block: bool argument to determine if residual block is used.
"""
super().__init__()
upsample_stride = upsample_kernel_size
self.transp_conv = get_conv_layer(
spatial_dims,
in_channels,
out_channels,
kernel_size=upsample_kernel_size,
stride=upsample_stride,
conv_only=True,
is_transposed=True,
)
if res_block:
self.conv_block = UnetResBlock(
spatial_dims,
out_channels + out_channels,
out_channels,
kernel_size=kernel_size,
stride=1,
norm_name=norm_name,
)
else:
self.conv_block = UnetBasicBlock( # type: ignore
spatial_dims,
out_channels + out_channels,
out_channels,
kernel_size=kernel_size,
stride=1,
norm_name=norm_name,
)
def forward(self, inp, skip):
# number of channels for skip should equals to out_channels
out = self.transp_conv(inp)
out = torch.cat((out, skip), dim=1)
out = self.conv_block(out)
return out
5.2.4 UnetOutBlock
class UnetOutBlock(nn.Module):
def __init__(
self, spatial_dims: int, in_channels: int, out_channels: int, dropout: tuple | str | float | None = None
):
super().__init__()
self.conv = get_conv_layer(
spatial_dims,
in_channels,
out_channels,
kernel_size=1,
stride=1,
dropout=dropout,
bias=True,
act=None,
norm=None,
conv_only=False,
)
def forward(self, inp):
return self.conv(inp)
到了这里,关于[论文笔记] Swin UNETR 论文笔记: MRI 图像脑肿瘤语义分割的文章就介绍完了。如果您还想了解更多内容,请在右上角搜索TOY模板网以前的文章或继续浏览下面的相关文章,希望大家以后多多支持TOY模板网!




![[深度学习论文笔记]UNETR: Transformers for 3D Medical Image Segmentation](https://imgs.yssmx.com/Uploads/2024/01/821708-1.png)